Section 1: Background, Abstract, and Data
| Background | |
|---|---|
| Gliomas are a type of tumor that occurs in glial cells in the brain or the spinal cord. The glial cells that are most prone to developing gliomas are astrocytes which are specific types of brain cells whose main function is to support neurons. [2] Gliomas that occur from astrocytes are called astrocytomas. Glioblastoma Multiforme, or GBM, is a type of glioma that exhibit more genetic abnormalities than other astrocytomas and are the most aggressive and fatal of gliomas. [3] Gliomas are categorized as high-grade and low-grade. GBM is high grade, and Low-Grade Gliomas (LGG) are low-grade. The causes of GBM are largely unknown, but research suggests that approximately 5 percent of GBM cases are caused by hereditary conditions, with some of the cases being from people with Neurofibromatosis type 1 (NF1), Turcot syndrome and Li-Fraumeni syndrome all being genetic syndromes associated with increased susceptibility to cancer. [4] Because there are multiple syndromes associated with GBM and hereditary causes comprise only a small percentage of the entire affected population, it is difficult to predict whether GBM will occur in an individual or not, and how dangerous they are in those harboring mutations. [5] | 
Source: https://www.mayoclinic.org/diseases-conditions/brain-tumor/symptoms-causes/syc-20350084 |
| Abstract | |
|---|---|
Source: https://www.mayoclinic.org/diseases-conditions/glioblastoma/cdc-20350148#dialogId52936835 |
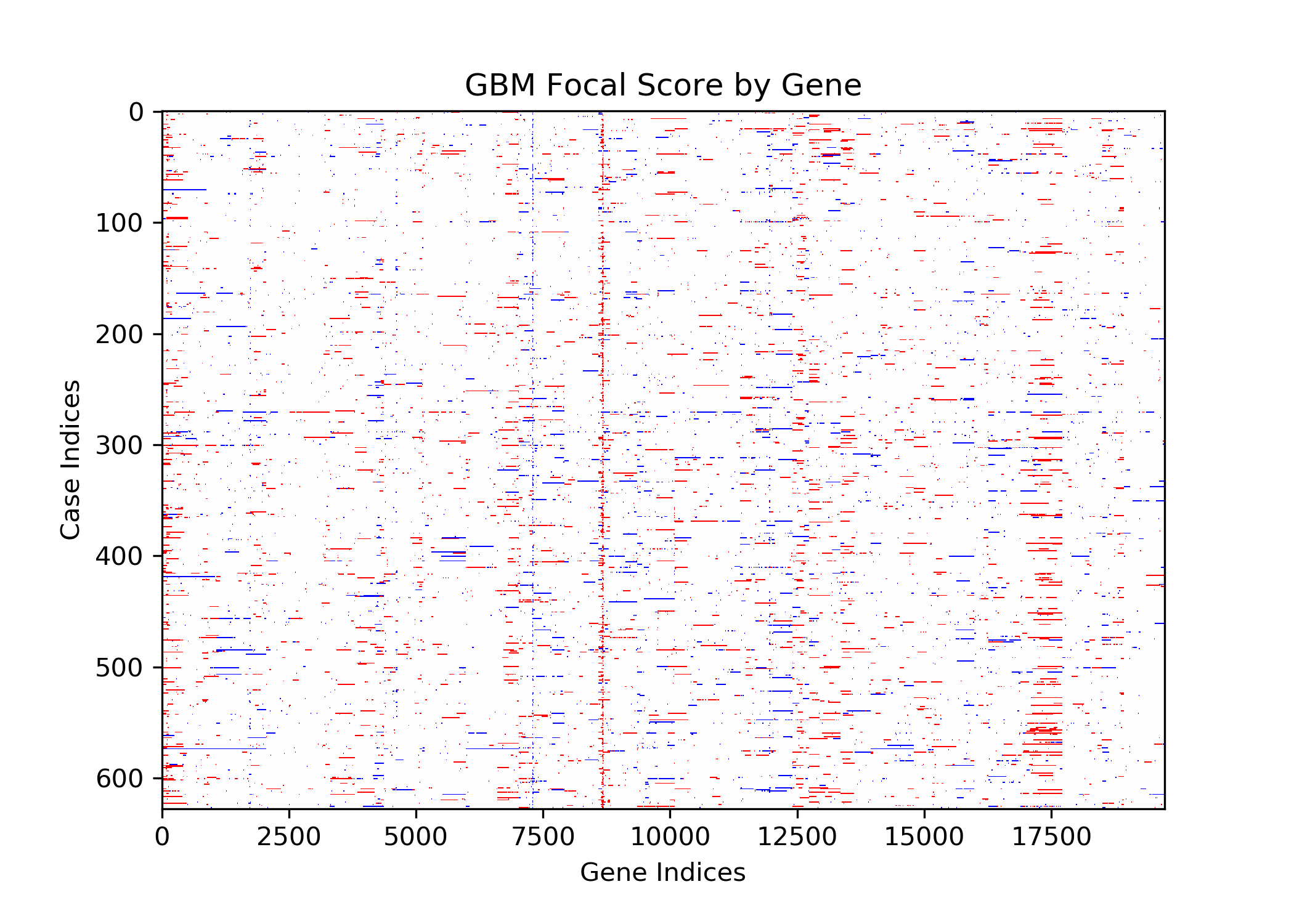
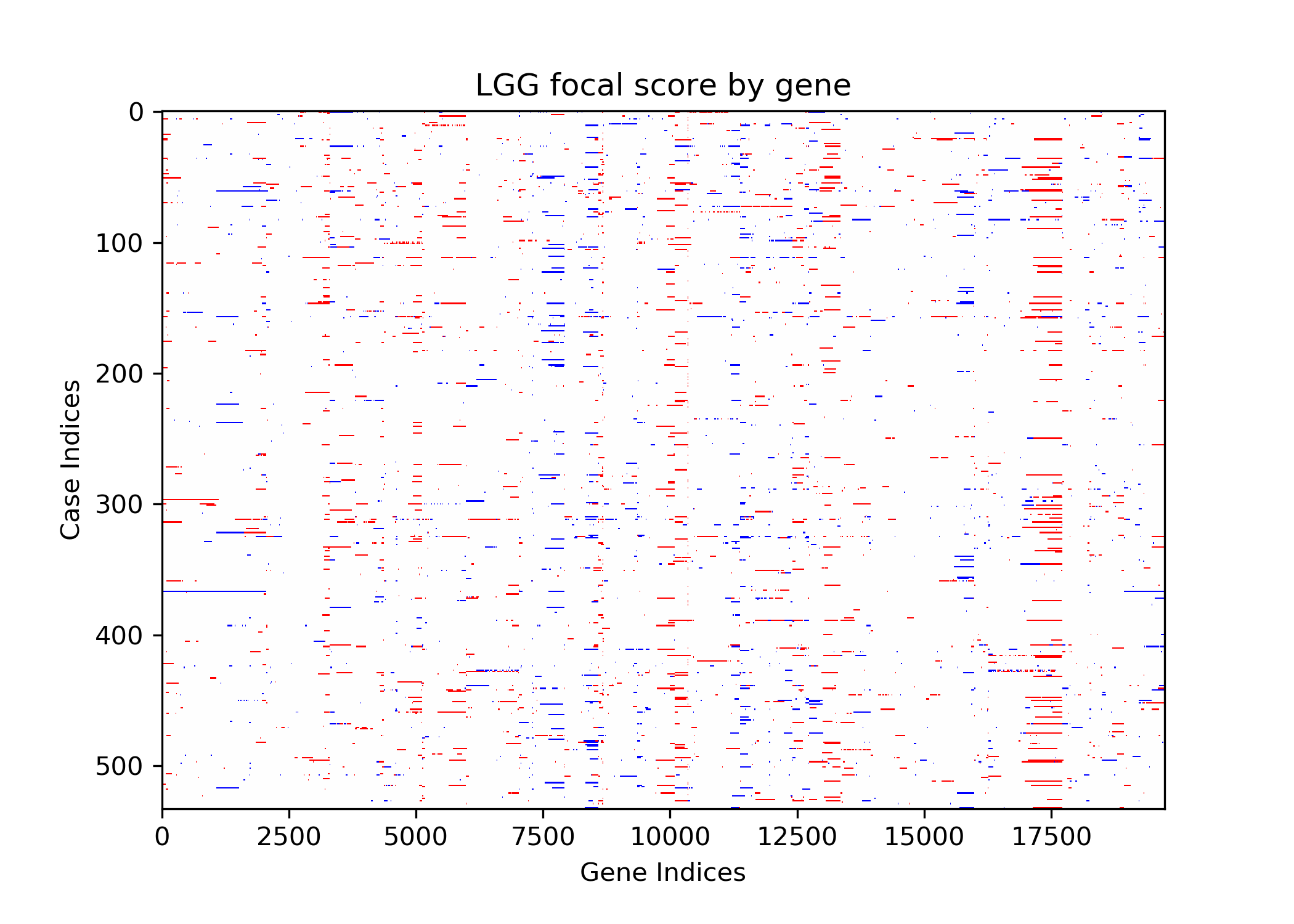
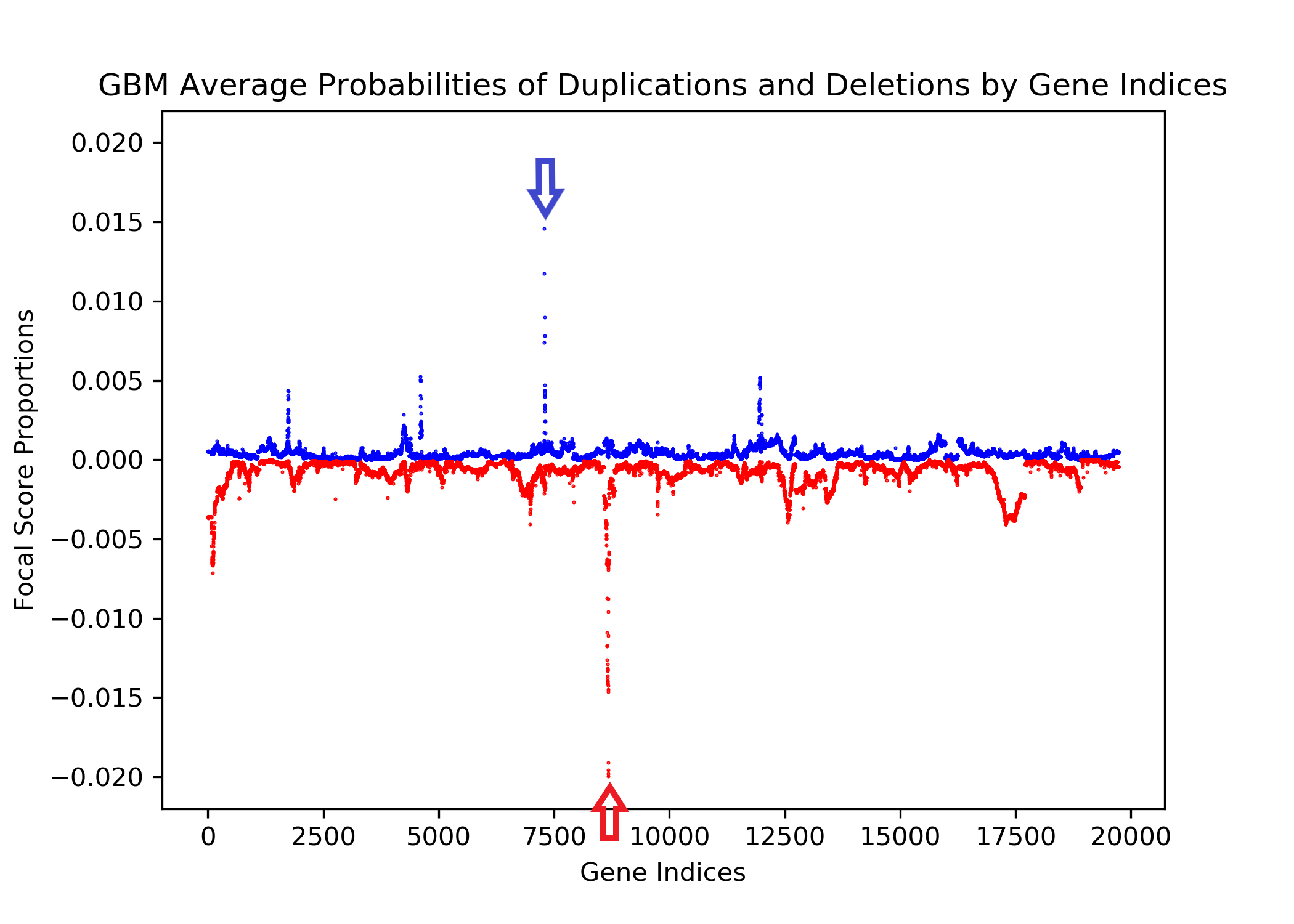
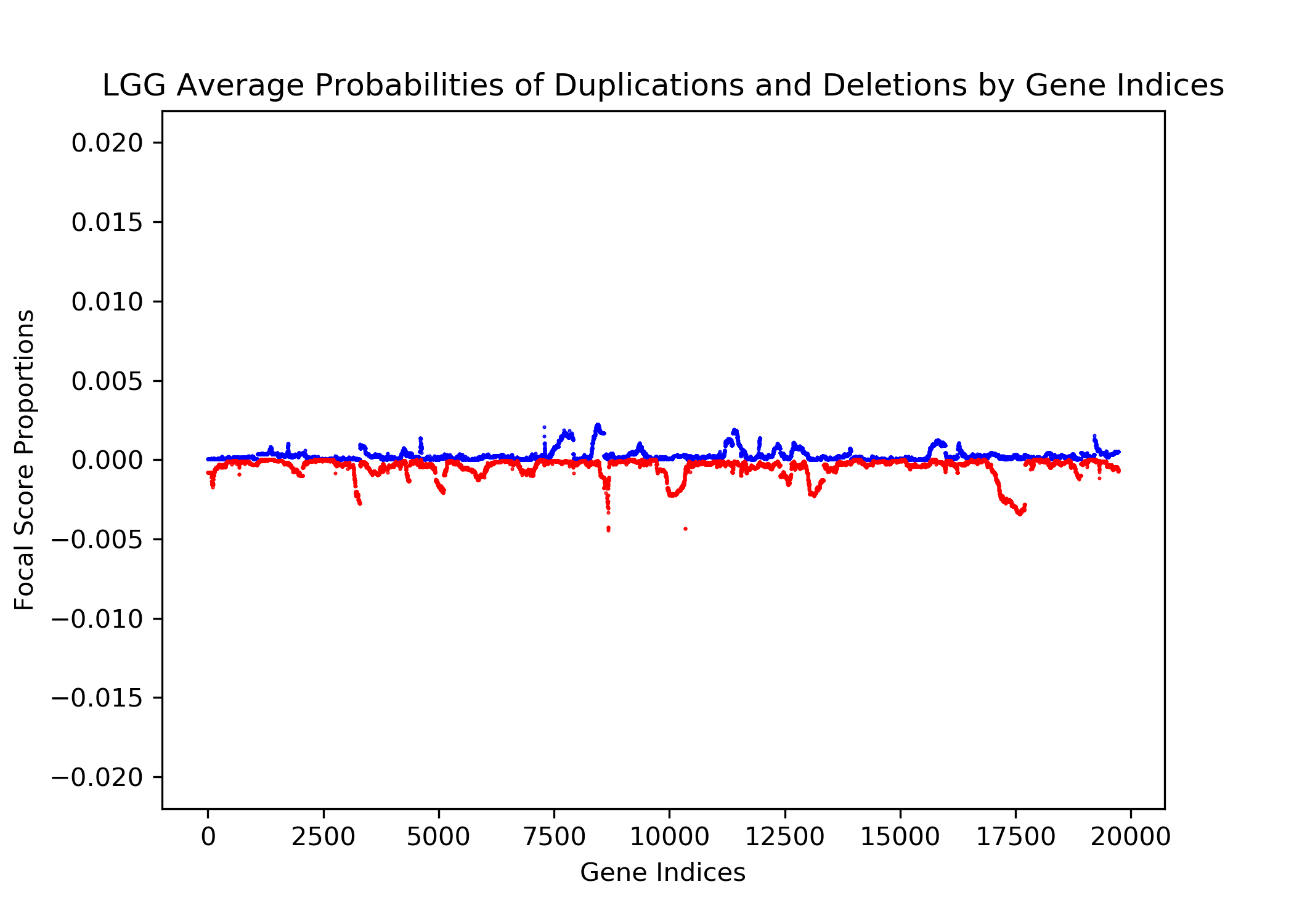
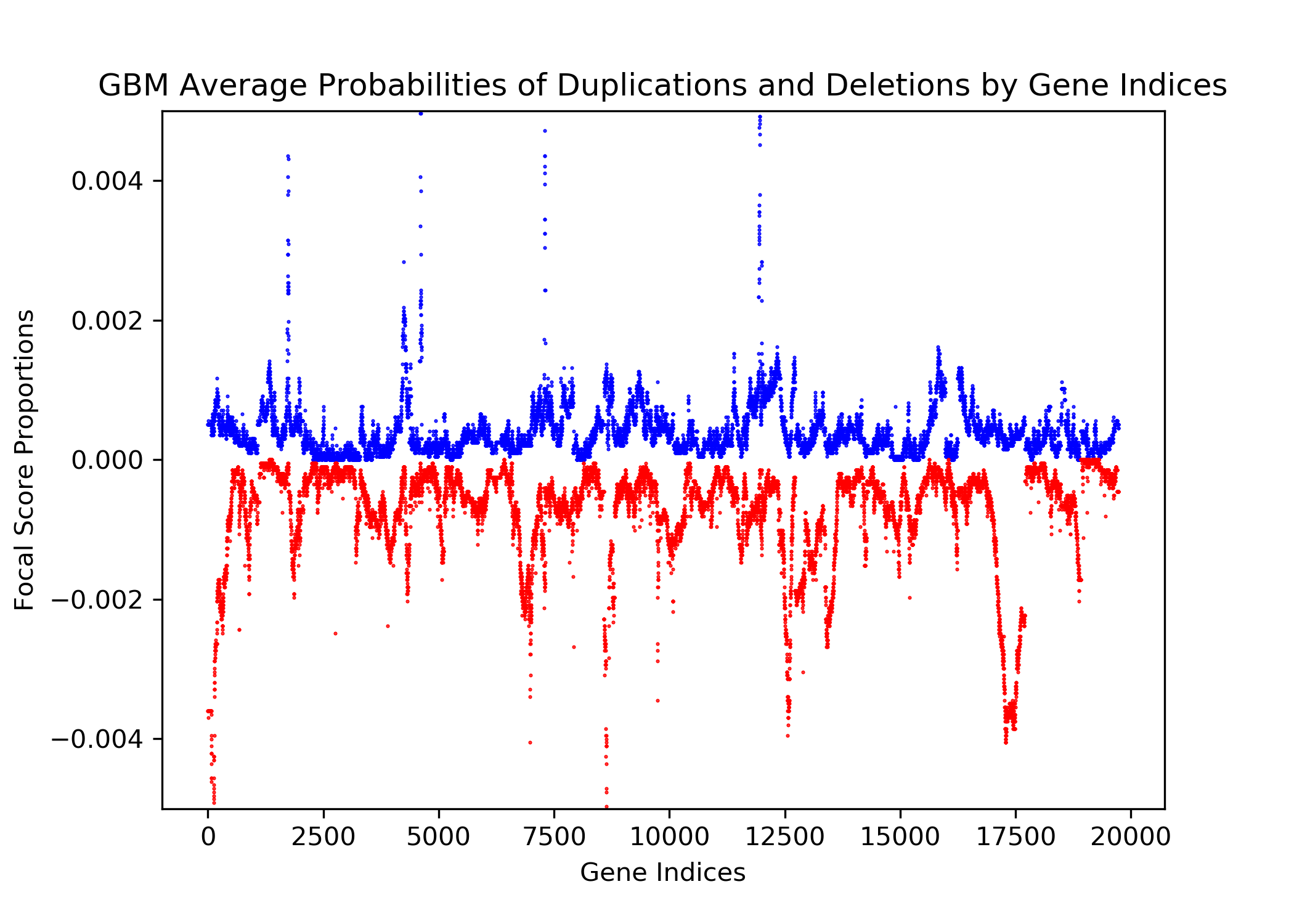
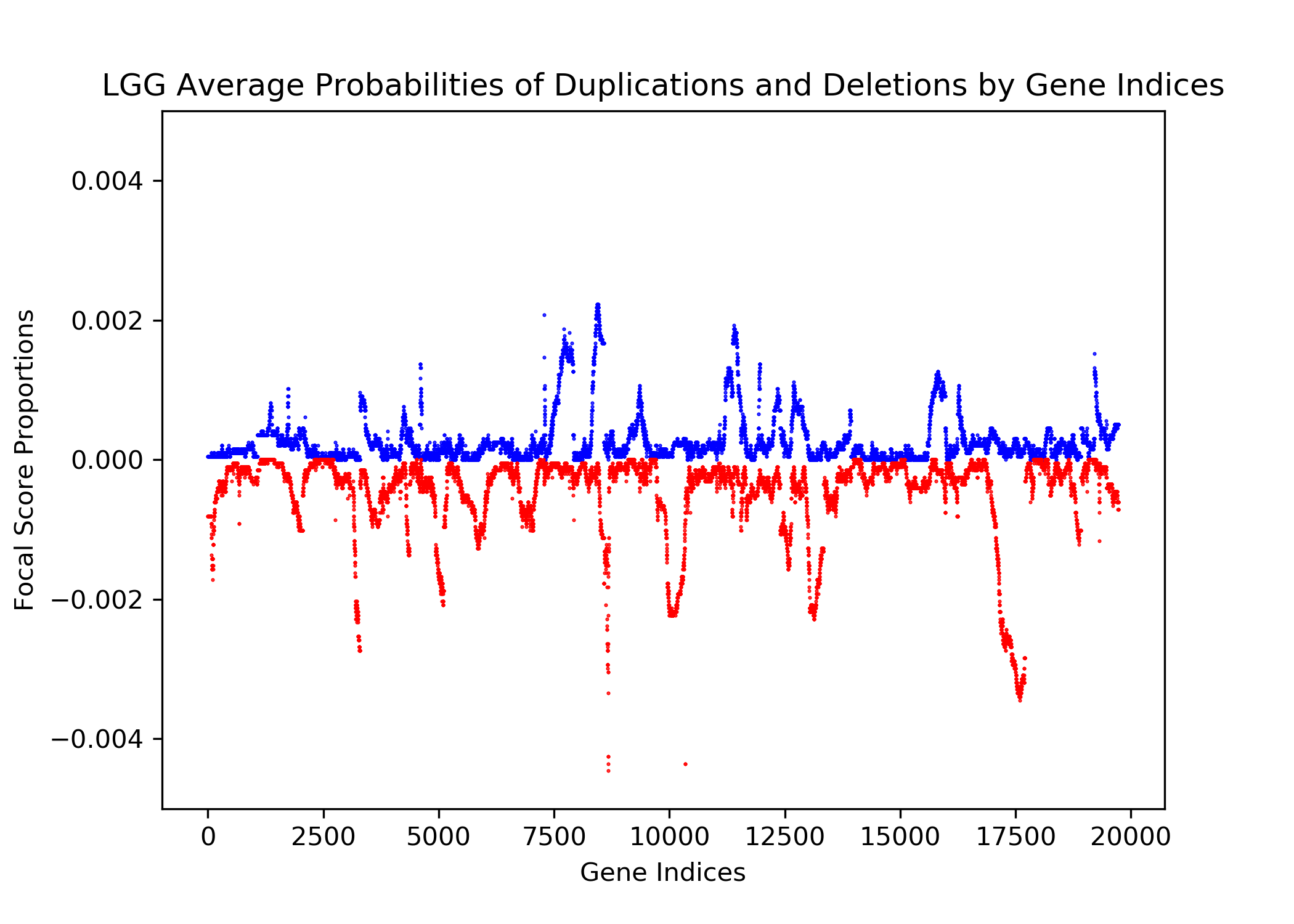
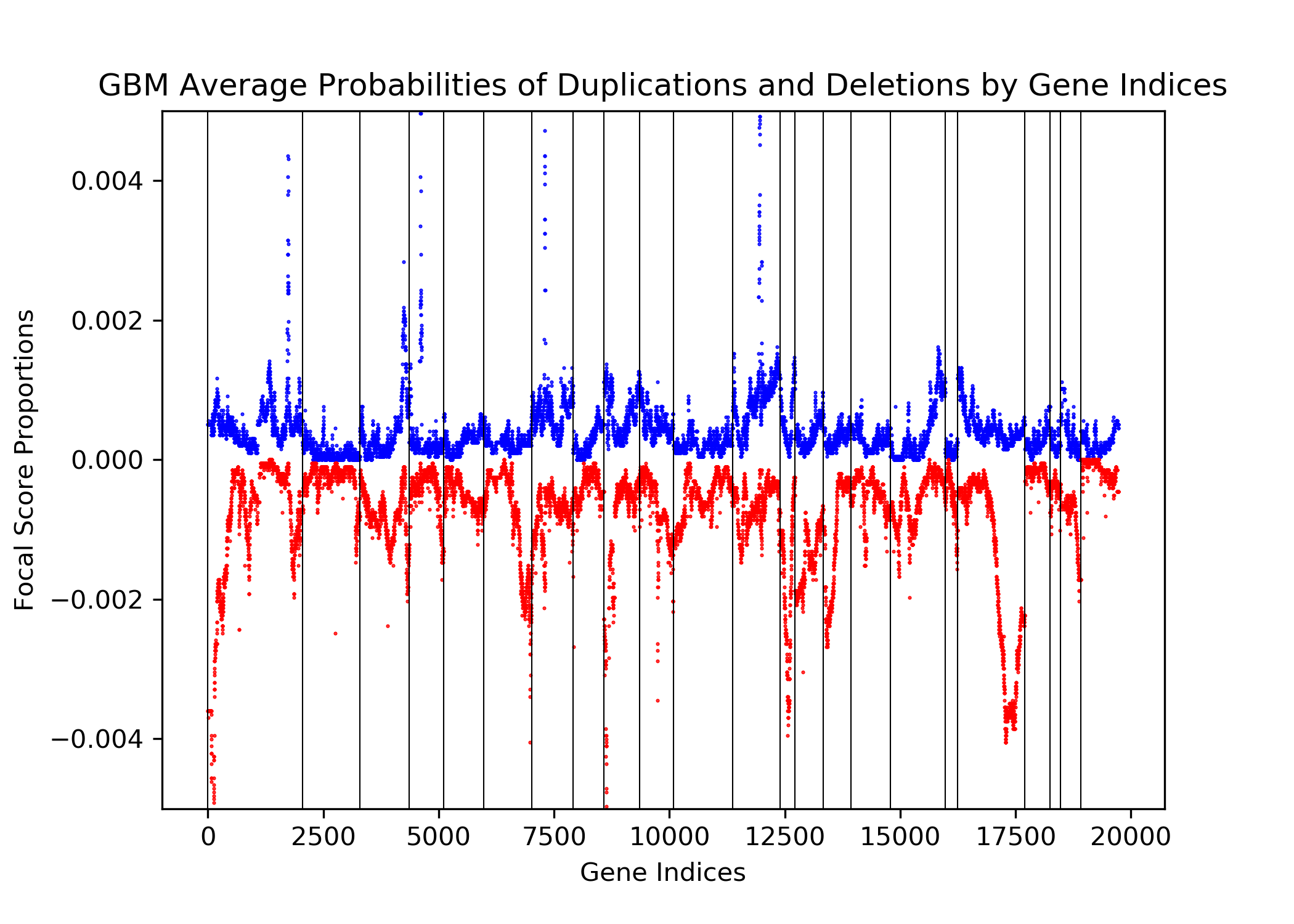
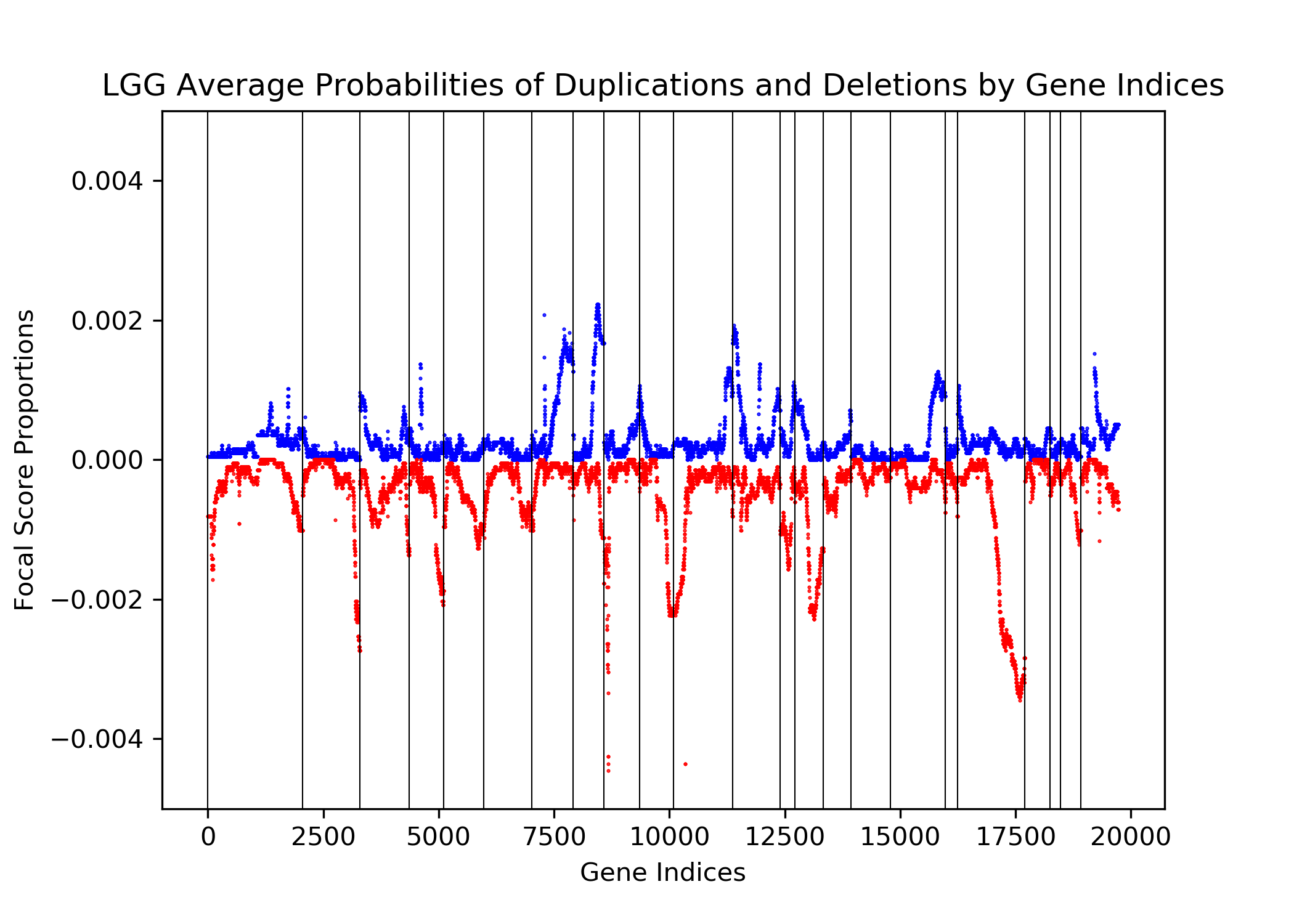
The goal of this project is to identify genetic differences between GBM and LGG populations. This is important because these genetic differences may be useful for diagnosing these diseases in people. This was done by comparing each population’s copy number variation focal scores. Our project’s question is: How does copy number variation of GBM and LGG distinguish? Our hypothesis is that the GBM population will have more copy number variation because GBM is more malignant, and so has more genetic mutations.In comparing the focal scores it was found that the GBM population had 1.5 times more duplications and deletions than the LGG population, confirming our hypothesis. And, there were specific sets of outlying genes in the GBM population that distinguish it from the LGG population - but only on a cross-case analysis level. Practically, it is not applicable to use them as markers for diagnosis. This project confirms the hypothesis that GBM has more genetic variation in its copy number variation, but does not support the approach to use copy number variation as a method of diagnosing either disease. We analyze the genetic differences by using the data from The Cancer Genome Atlas (TCGA) which is an online repository of cancer data. To understand how we analyze the genetic differences, we would introduce some important terminology that are helpful. |
| Terminology | |
| Copy Number Variation | The variation of numbers of copies of a gene. This variation occurs from mutations and are categorized as duplications, deletions, or segmental duplication. Cancer cells are typically related to a set of multiple mutated genes, so copy number variation is a good indicator for some diseases like cancers. |
| Focal Scores | Summarize copy number variation. In our data, a score of one indicates a gene duplication, a score of zero indicates that there was no change in copy number, and a score of negative one indicates a gene deletion. |
| Data Overview | |
|---|---|
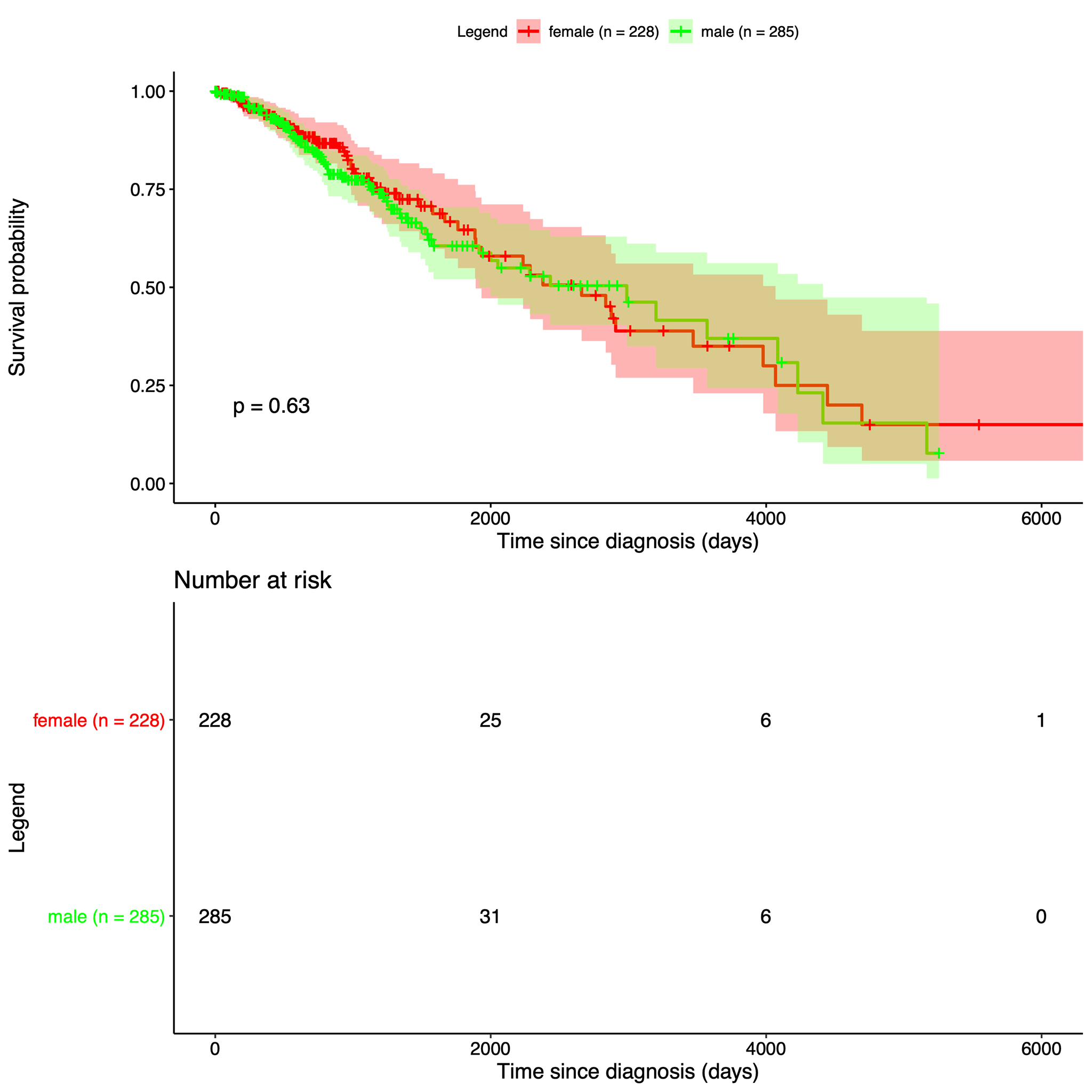
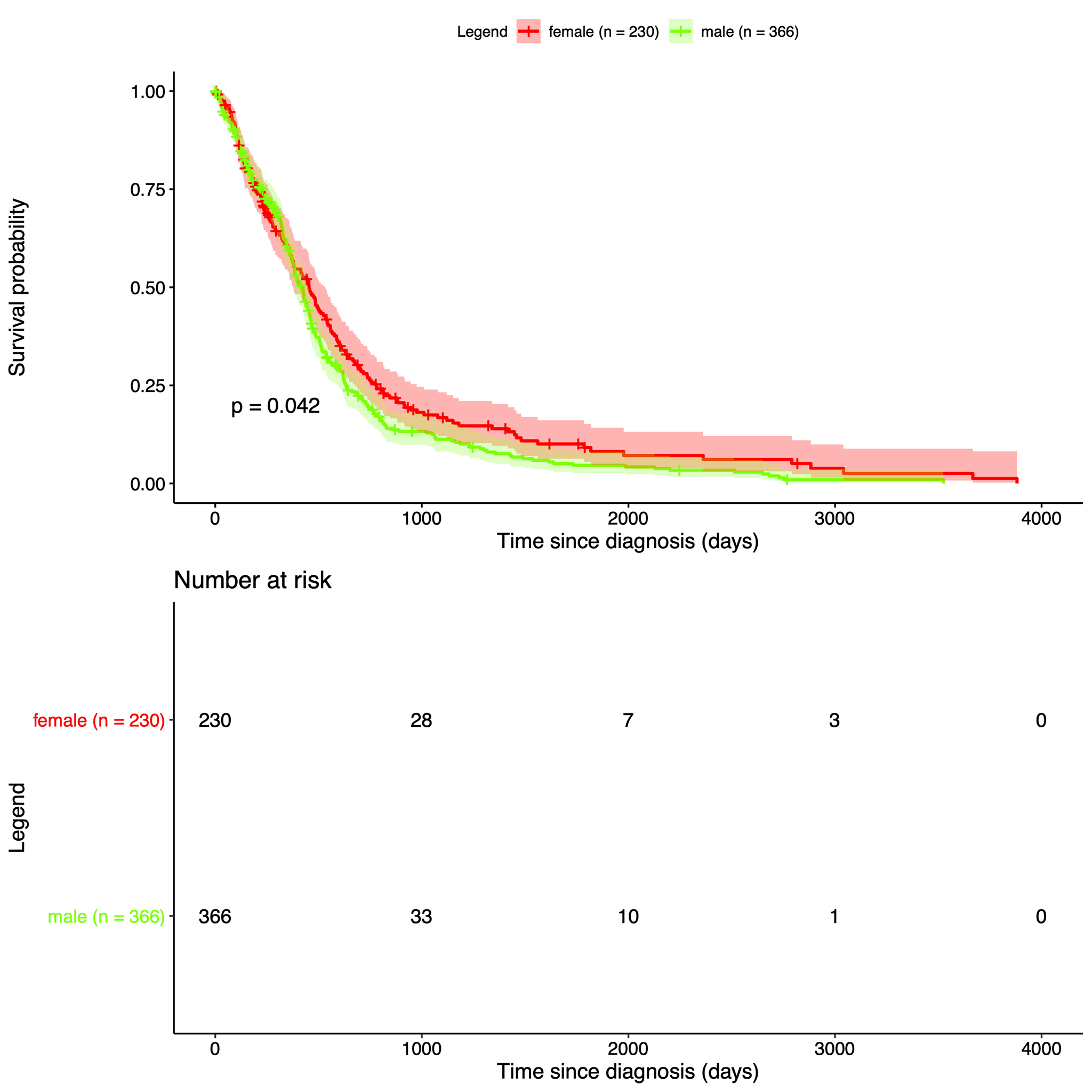
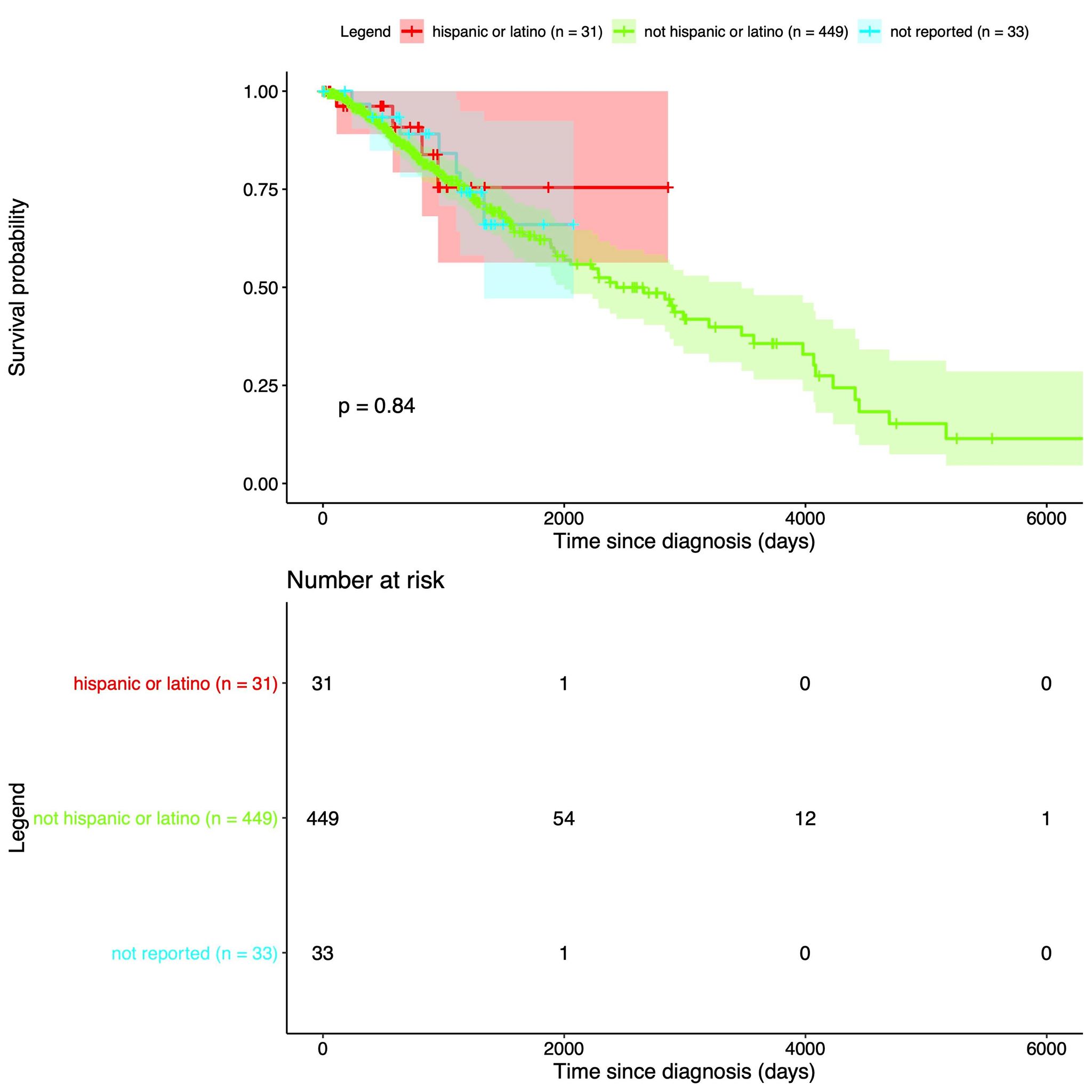
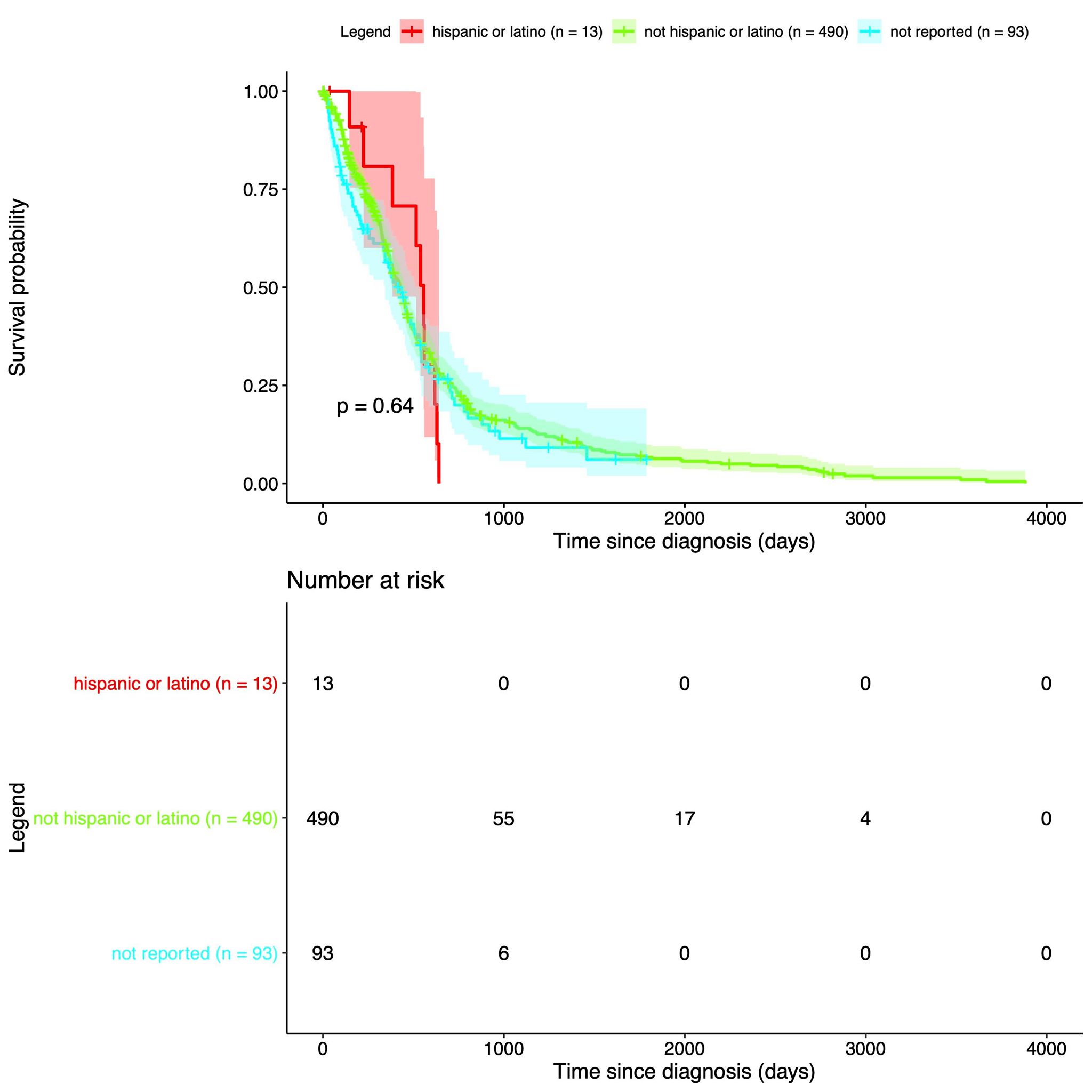
| Our data is from The Cancer Genome Atlas, which is an online repository of cancer data. It was started by a diverse group of researchers to consolidate and analyze cancer data. Their data portal is called the Genomic Data Commons Data Portal, or, GDC. The GDC has over 84,000 cancer cases spanning across 67 primary cancer sites such as the lung, breast, or brain. In our project we used all of their Glioblastoma Multiforme cases and all of their Low-Grade Glioma cases, which amounted to 617 and 507 cases respectively. In those cases they have both CNV data and clinical data. The clinical data includes gender, when they were diagnosed, and when they passed away, if they passed away. | 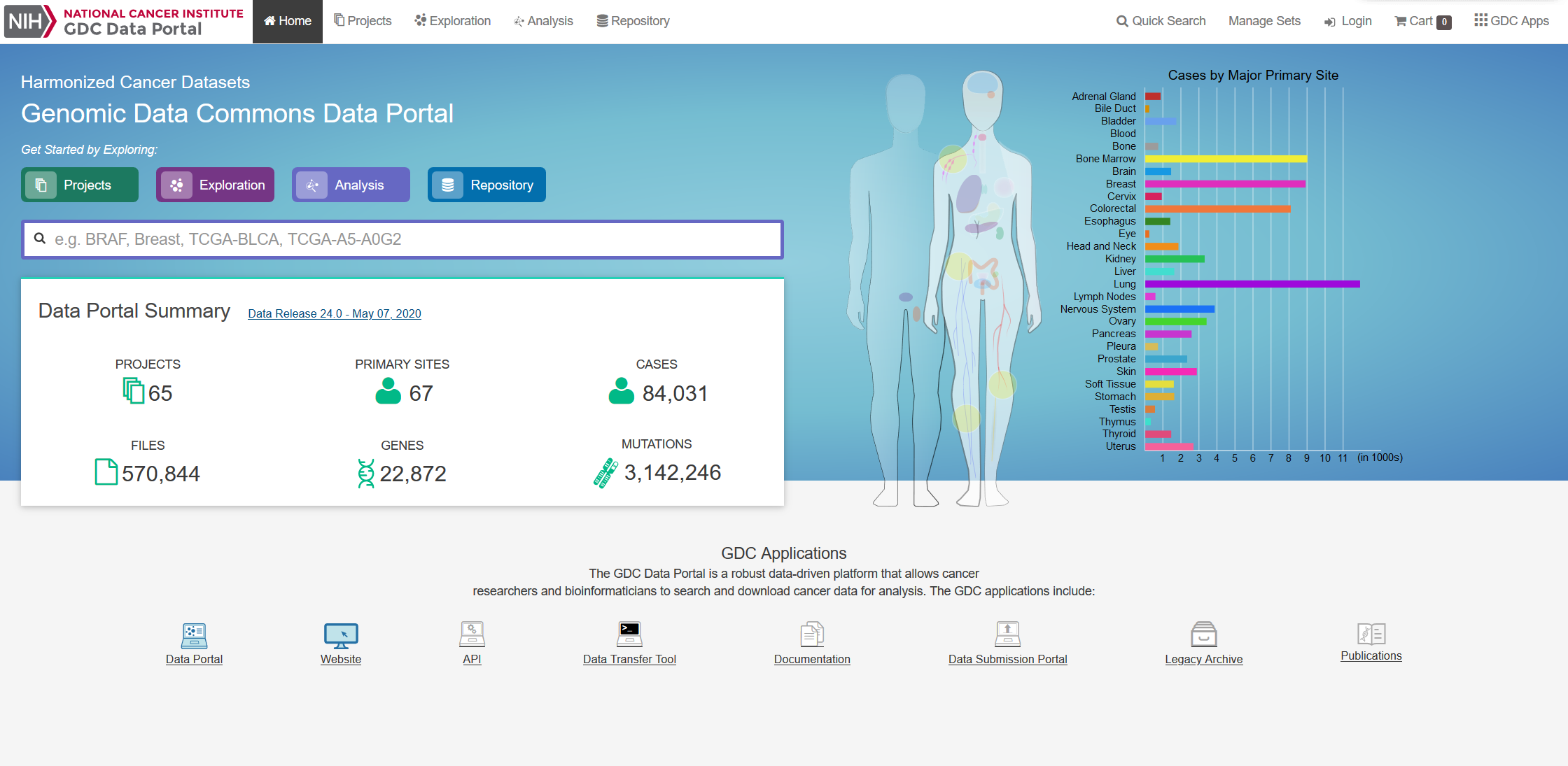
Source: https://portal.gdc.cancer.gov/ |
| Data Ingestion | |
|---|---|
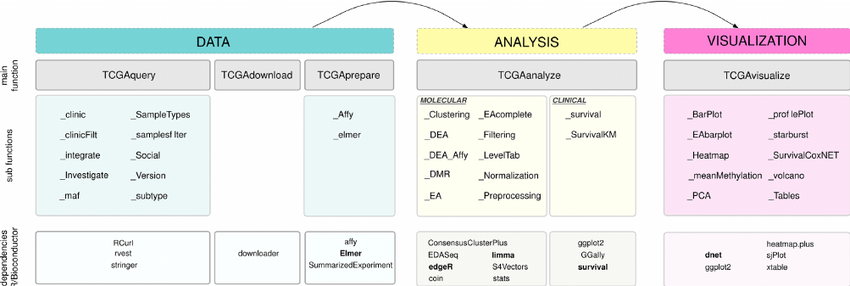
Source: https://bioconductor.org/packages/release/bioc/html/TCGAbiolinks.html |
Our data ingestion pipeline consists of three modules: Data, Analysis, and Visualization. Under the Data Module, the filtered data will be downloaded and prepared for the model to digest. In the Analysis Module, the pre-processed data will be fit into our models. Our clinical data was ingested automatically with an R package called TCGAbiolinks. To gather the data, all we need to do is to set a query similar to a SQL query. For our Copy Number Variation, all of the data we needed was stored in one text file, which could just be downloaded directly from the GDC. |